Our research
We focus on developing robust artificial intelligence (AI)-powered multimodal risk-stratification tools to forecast the risk of disease progression in cancer patients. These tools incorporate the composition of the tumor microenvironment (TME) in their training to improve their robustness, accuracy, and generalizability to different patient cohorts. We work with different data modalities, including transcriptomics, histopathology, and radiology images to build robust AI models for risk assessment in patients newly diagnosed with cancer. A key component of our research is using the composition of the tumor microenvironment to extract high-quality biological features that serve as supervisors for the training process, guarding against overfitting.
Our work focuses mainly on prostate cancer; however, it is important to note that our approaches can be applied to other cancer types as well
Identifying digital pathology signatures of high-risk cancer phenotypes
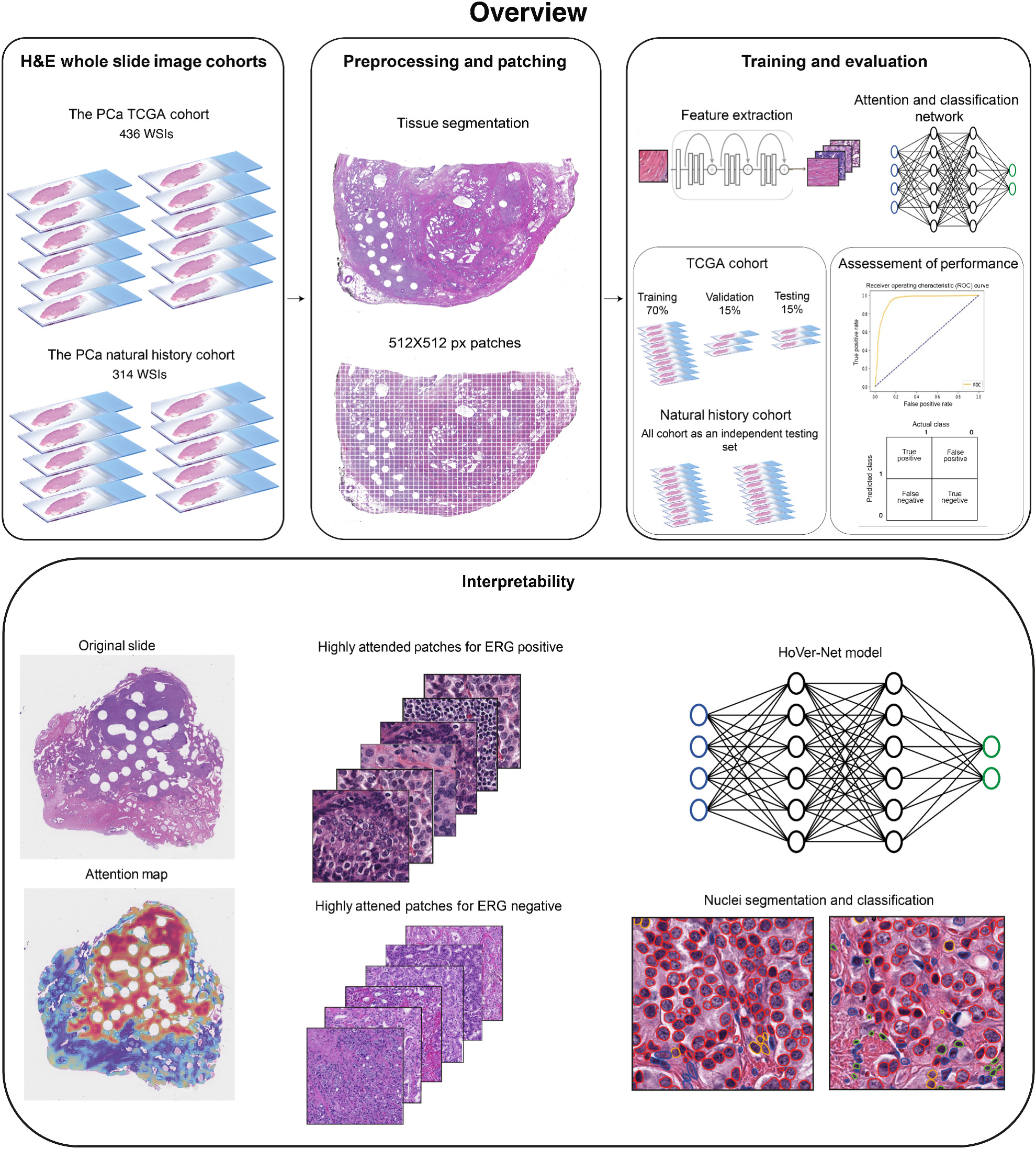
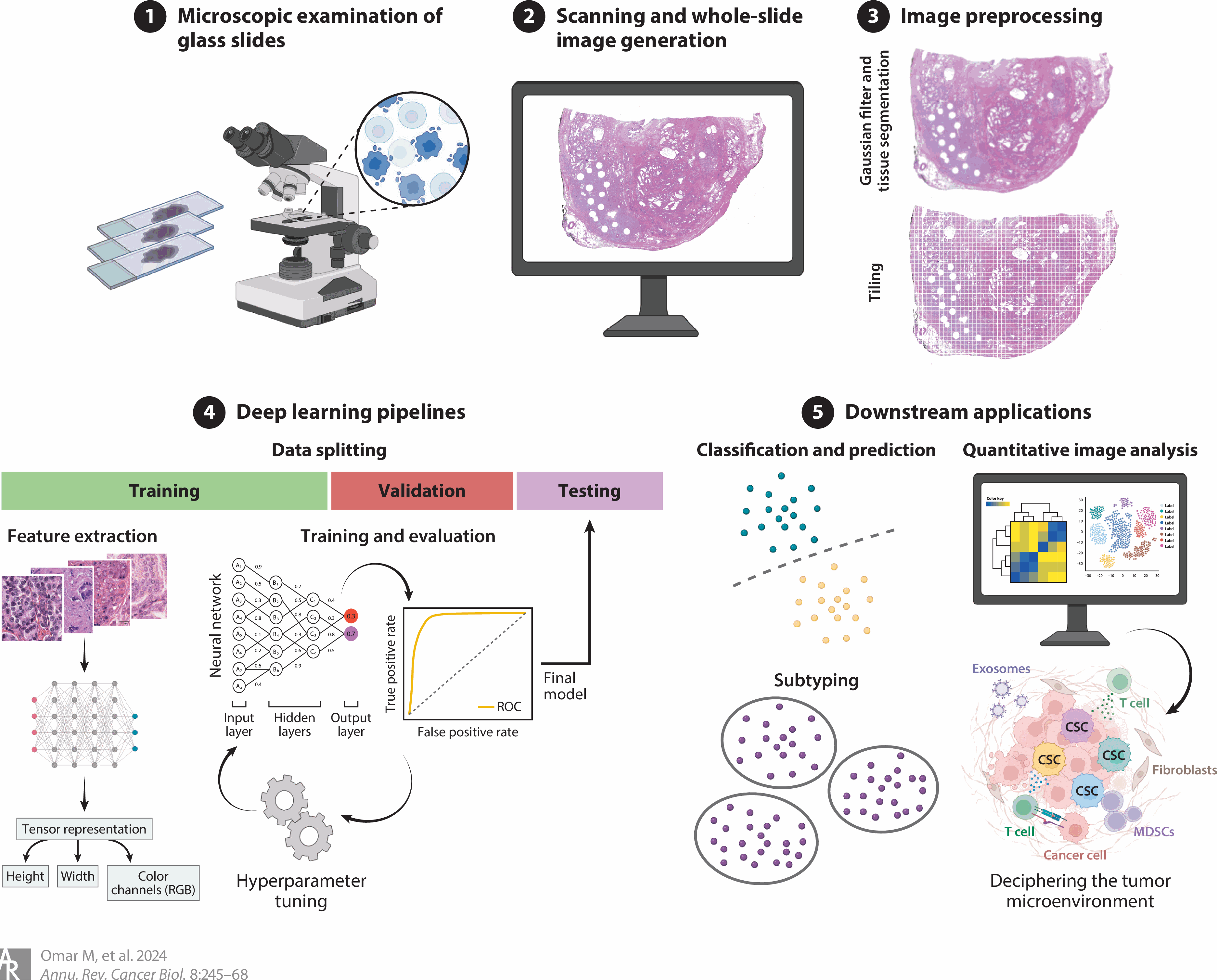
We are interested in uncovering pathomic signatures from routine H&E-stained whole slide images (WSIs) to inform patients’ prognostication and the prediction of molecular and clinical phenotypes (see Omar et al. Ann Rev of Cancer Bio. 2024). Our work in this domain leverages deep learning algorithms to automate WSIs preprocessing and feature extraction to identify morphometric features associated with certain phenotypes. For instance, we recently developed a robust model for inferring the status of TMPRSS2:ERG fusion (a key molecular alteration in prostate cancer) from the tissue morphology depicted in routine H&E-stained images of radical prostatectomy specimens (see Omar et al. Mol Cancer Res 2024).
Deconvoluting the tumor microenvironment composition across the spectrum of cancer initiation and progression
Our lab focuses on developing stage-specific tissue modules that comprehensively capture the complex composition and interplay within the tumor microenvironment at early stages preceding progression. These modules comprise cellular composition, expression profiles, intercellular interactions, gene regulatory networks, and spatial neighborhoods. This approach provides an in-depth, systemic understanding of the tumor microenvironment, highlighting the diversity of cell types and the extensive network of molecular and spatial interactions that dictate cellular function and the trajectory of progression. We employ high-resolution spatial omics to generate single cell-resolution data that recapitulates the spatial distribution and abundance of mRNAs and specific proteins within the tissue context. We leverage this data to construct detailed maps of cellular composition, molecular expression and spatial localization, facilitating a nuanced analysis of how these factors interact within the physical confines of the tumor microenvironment as the tumor progresses.
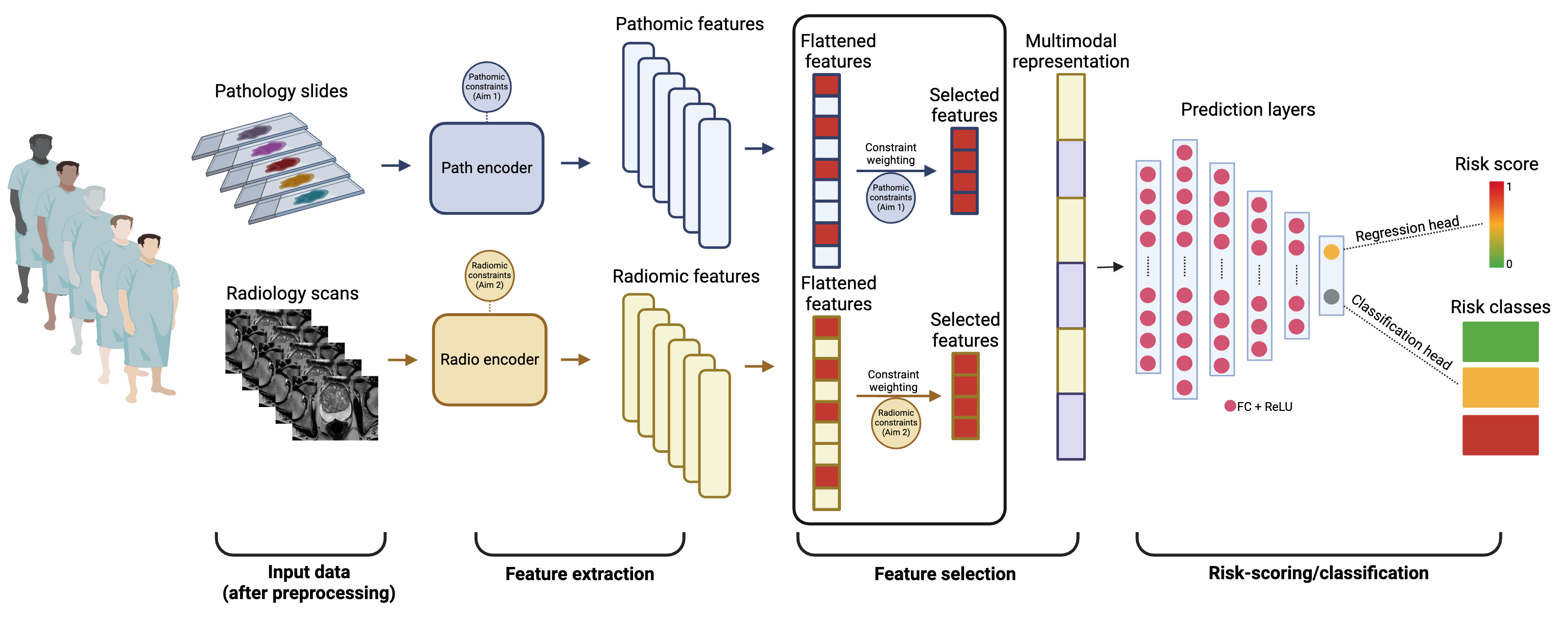
Developing multimodal risk stratification tools for cancer patients to assist with clinical decision making
Our lab develops multimodal risk stratification tools that integrate diverse patient-centered data types, including omics, pathomics, and radiomics to enhance risk assessment and inform patients’ management. Omics offer a deep dive into the molecular underpinnings of tumor progression, while pathomics provide a spatial microscopic view of TME dynamics by translating pathology images into quantifiable data. Radiomics further enrich this by extracting non-invasive macroscopic features from medical imaging that can be correlated with underlying disease mechanisms and outcomes. The fusion of these data modalities promises to unveil intricate indicators of high-risk disease at very early stages, which cannot be discerned using unimodal approaches.